Our lab harnesses root developmental plasticity to study how post-embryonic tissues and organs are initiated in different developmental and environmental contexts. Our long-term goal is to define fundamental principles underlying the specification and function of regenerative cells. Taking advantage of the simplifying properties of Arabidopsis, a genetically tractable model plant, we apply cutting edge omics and imaging techniques to investigate cell fate decisions and link them to cell behavior.
Research Directions
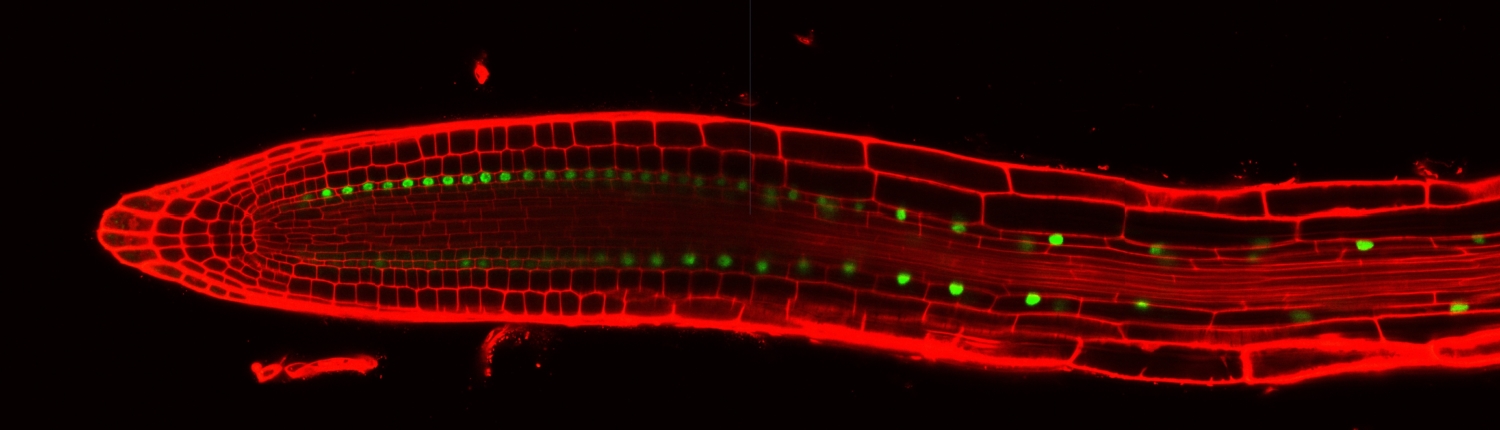
Elaboration of the plant body plan. As shown in the video above, the Arabidopsis root system is elaborated by the periodic formation of lateral roots which emerge horizontally from the vertical primary root. The spatiotemporal patterning indicates that only some cells in the primary root will initiate a lateral root. A key question is what determines this subset of cells.
Previous work found that a luciferase reporter driven by a synthetic promoter, DR5, is rhythmically expressed and marks the primary root cells that will eventually form a lateral root. Microarray experiments of finely dissected primary root sections identified genes that oscillate in phase and anti-phase with the DR5:Luciferase reporter. This observation led to the hypothesis that a biological oscillator specifies the primary root cells that will become ‘founder cells’ and later divide to form the new stem cell niche from which a lateral root will emerge. It is unknown what transcriptional mechanism regulates the oscillations and how one or multiple cell types remember and respond to this transient event.
Using lateral root development as a model, our lab applies single cell omics approaches to investigate how gene regulatory programs in distinct primary root cell types orchestrate organ neogenesis.
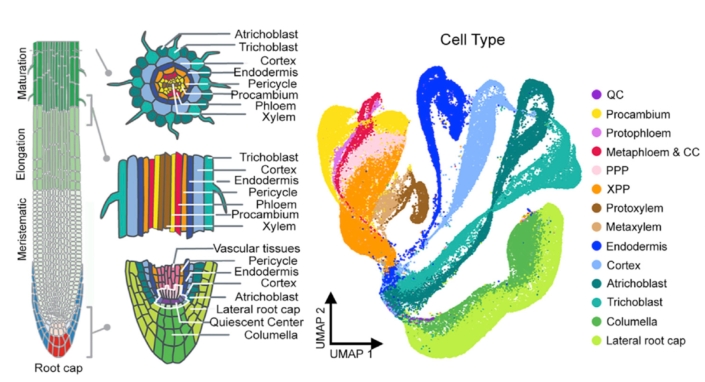
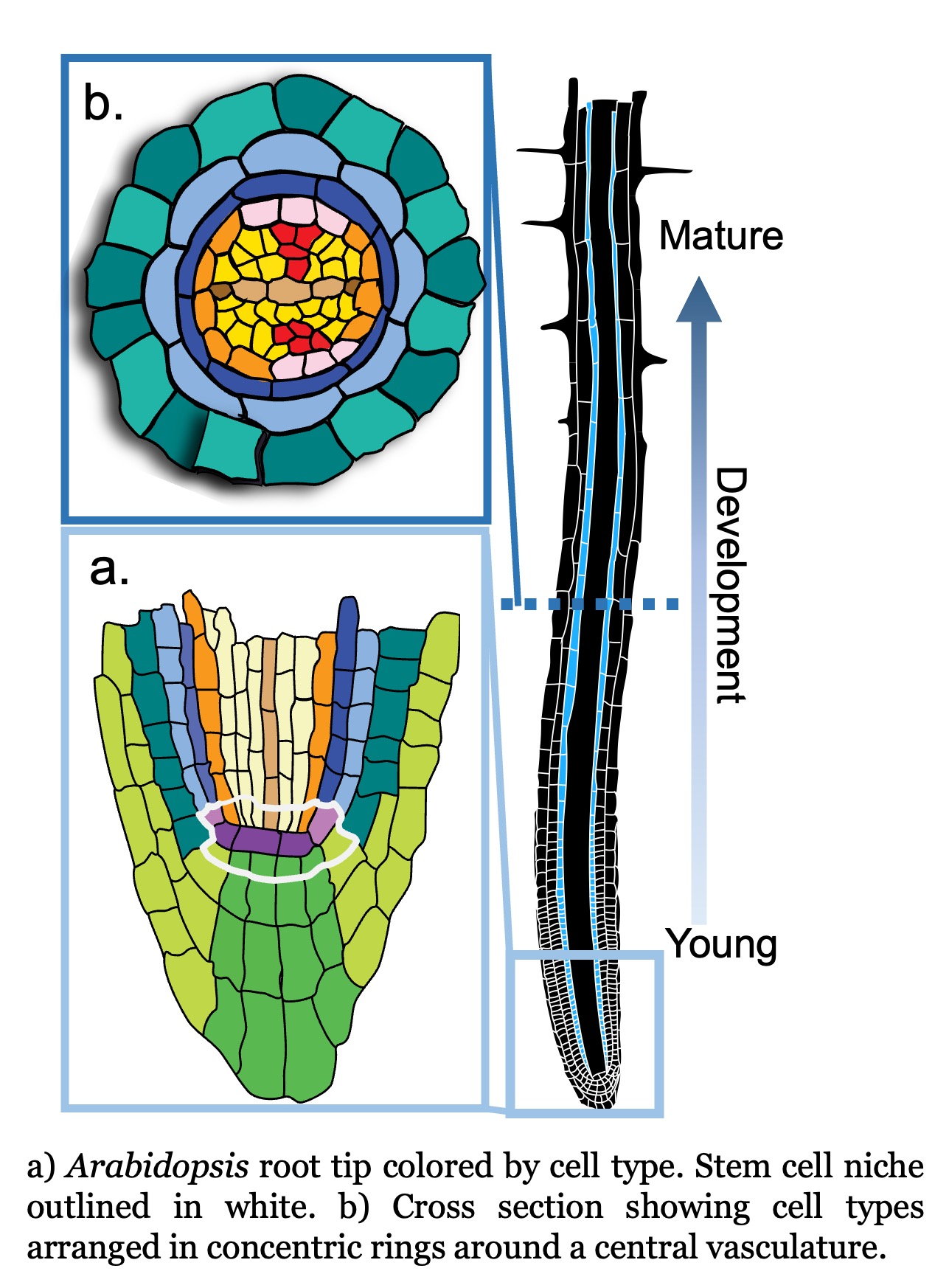
Age-related developmental changes. Development happens in four dimensions where the fourth dimension is time. The Arabidopsis root, with its radial symmetry and immobile cells, simplifies a 4D problem to 2D. Thus, the root enables us to study cell maturation, a continuous developmental process, with information collected from a snapshot in time.
Using single cell RNA-seq data from a single snapshot, we ask questions such as i) how does gene expression change over developmental time; ii) what transcription factors drive cell fate decisions; and iii) how do genetic perturbations affect cell identity specification and maturation? We are now poised to ask how gene expression changes over actual time as a seedling ages.
We generated time series single cell RNA-seq data from root tissue collected at 24 hour intervals. We are asking how age-related molecular changes in specific cell types relate to changes in root tissue patterning and root system architecture. We are specifically interested in understanding:
i) If there are age-dependent transcription factors and gene regulatory networks controlling cell fate specification
ii) If there are age-related differences in the connections between cell fate and cell division decisions