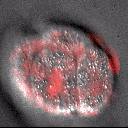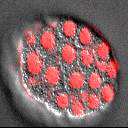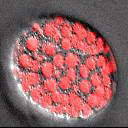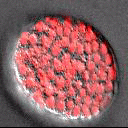
A part of the Molecular, Cell, and Developmental Biology Department (MCDB) at UCLA and the Institute for Quantitative and Computational Biosciences (QCBio), we apply advanced imaging and computational approaches to understanding organismal development.
The Development of Behavior
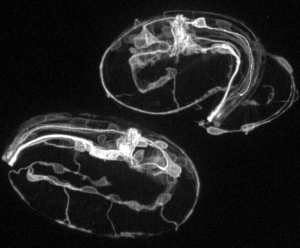
We are exploring these questions using the embryo of the nematode Caenorhabditis elegans as a model. With a combination of methods drawn from classical genetics, the state-of-the-art in quantitative imaging, and new approaches to manipulating single cells in vivo, we are mapping the emergence of functional circuitry in the embryo. While cell adhesion molecules and morphogen gradients may teach us how to ‘wire’ a brain, we are working to understand how to ‘boot’ one up.
Temporal Patterns and the Regulation of Cell Fate
The regulation of cell cycle progression is a central problem for many fields of biology. We’re interested in understanding how the length of the cell cycle itself acts as a regulatory input into developmental decisions, such as the fate decisions made by stem cells in the early embryo. We combine classical genetic perturbations with light microscopy and environmental manipulations such as precise temperature control to probe the mechanistic links between cell cycle and cell fate regulation.
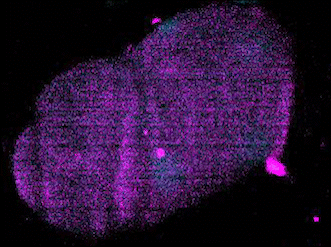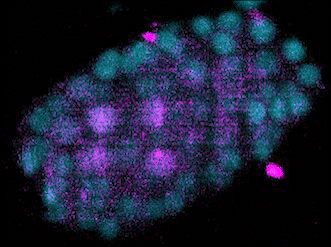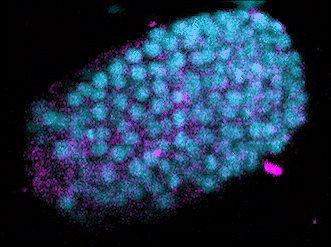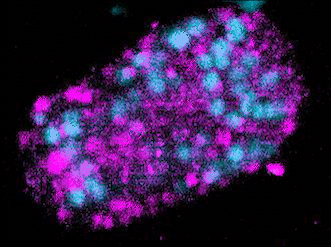
The Evolution of Development
The broad availability of natural diversity and multi-species resources among nematodes has made them an attractive system for the study of the evolution of development. Using machine learning and spatial transcriptomics approaches, we are working to build developmental and molecular maps of fate specification across diverse nematode species and to identify mechanisms by which natural genetic variation builds towards species-defining shifts in developmental strategies. Key to this is our work developing automated label-free approaches to tracing cell lineages in nematode strains and species for which transgenesis is either not currently possible or a major bottleneck for research.